| Home |
| The Team |
| Lab chronicle |
| Open Position(s) |
| Teaching |
| Research |
| Collaborations |
| Publications |
| Funding |
| Disclaimer |
 |   | |
Unit of Plant Molecular Cell Biology | ||
Open Position(s) We regularly look for talented candidates for bachelor´s and master´s theses. Please contact us for further information. We currently seek for a
talented PhD student to pursue a DFG-funded project at the cross-junction
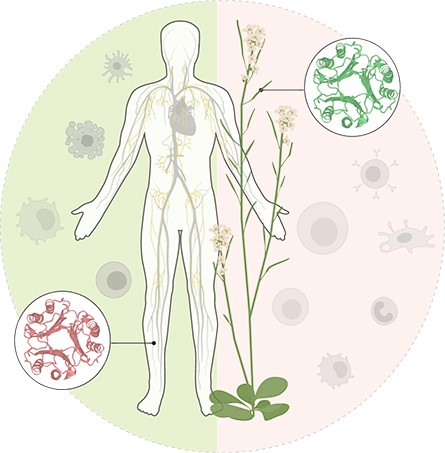
of plant biology and human immunology. The project deals with the functional
analysis of Macrophage Migration Inhibitory Factor/D-Dopachrome Tautomerase
(MIF/D-DT)-like (MDL) plant proteins. These MDL proteins are orthologs of human
MIF, which is a chemokine and key regulator of human immunomodulatory
activities. Plant MDL proteins are poorly characterized to date, and in tight collaboration
with the Bernhagen lab at the LMU Munich
we aim to study their functions using a cross-kingdom approach (see references
below). The PhD position is funded for three years at the level of a TVöD 65%
E13 position. We expect a Master´s degree in biology or a related subject, good
knowledge in (plant) genetics, molecular biology and biochemistry, and the
willingness to engage actively in the collaborative project across universities.
Please send your application (letter of motivation, CV, and certificates) to panstruga@bio1.rwth-aachen.de
by 31st March 2025 at the very latest.
collaboration-to-study-host-immunity-in-plants-a Key references: Spiller et al. 2023. Plant MDL proteins synergize with the cytokine MIF at
CXCR2 and CXCR4 receptors in human cells. Science
Signaling 16: eadg2621 Gruner et al. 2021. Chemokine-like MDL proteins modulate flowering time
and innate immunity in plants. Journal
of Biological Chemistry 296: 100611. Sinitski et al. 2020. Cross-kingdom mimicry of the receptor signaling and leukocyte recruitment activity of a human cytokine by its plant orthologs. Journal of Biological Chemistry 295: 850-867. We also currently offer a very attractive Master’s thesis about AI-based simulation and structure-informed verification of protein complex interfaces under the supervision of Dr. Jan Hübbers (starting date: January 2026 or subject to negotiation) . Our group investigates the interaction between model and crop plants and their phytopathogenic powdery mildew fungi. We study various cellular and molecular processes underlying these interactions. A particular focus of our research are pathways that establish cell polarity and may play key roles in plant–powdery mildew interactions. Understanding the complex network of proteins involved in these processes is tedious. Thus, we use modern AI tools for protein complex modeling that can predict protein–protein interfaces and thereby guide experimental validation. We have established a functional pipeline for AI-based protein interface prediction and are now looking for enthusiastic students to apply and further develop this cutting-edge approach. This Master’s project offers a unique opportunity to combine AI-driven structural biology with experimental (wet-lab) research and thus provides an excellent entry point into an emerging field. Your tasks • Apply our AI-based modeling pipeline to identify potential protein complexes in silico. • Use downstream analytical metrics and visualization tools (e.g. ChimeraX) to pinpoint interface residues critical for protein complex formation. • Perform in silico mutagenesis of these residues to assess their role in complex stability and disruption. • Experimentally verify complex formation and disruption through in vitro targeted mutagenesis and in planta luciferase complementation assays. Your profile • Bachelor’s degree (or equivalent) in biology, biotechnology, or a related field in the natural or computational sciences. • Strong interest in combining experimental and computational approaches. • Basic knowledge of programming languages such as Python or R is not required but considered a plus. • Good written and spoken English communication skills. We offer • A multidisciplinary research environment that bridges fungal and plant cell biology and computational modeling. • Hands-on training in AI-based structural analysis and experimental validation techniques. • Comprehensive onboarding, daily exchange with your supervisor, and feedback from the group leader in regular lab meetings. • A small lab with a supportive and collaborative culture that encourages initiative and scientific growth. Contact and application Please send (i) your CV and (ii) an overview of grades from your Bachelor’s and Master’s studies ideally by 2025-11-30. For questions or to apply, contact Dr. Jan Hübbers: jan.huebbers@rwth-aachen.de. | ||
| Last updated: 25.10.2025 | ||