Research overview
Our research activities are mainly devoted to study cell biological
phenomena in the context of plant-microbe interactions. In particular,
we focus on the interactions between plants and powdery mildew fungi,
obligate biotrophic ascomycetes that cause agronomically relevant
diseases (Figure
1).
One of our major interests is to unravel the cellular mechanisms of
plant defence, but we are also interested in the molecular details of
fungal pathogenicity.
Our lab employs a mixture of molecular biology, cell biology,
biochemistry, plant genetics and fungal genomics to pursue these goals.
The main plant species used in our research projects are the
dicotyledonous reference species, Arabidopsis thaliana, and the
monocotyledonous cereal, barley (Hordeum vulgare). Accordingly,
on the fungal side emphasis is given to powdery mildew species
colonizing barley (Blumeria hordei) and Arabidopsis (Erysiphe
cruciferarum).
Current research projects comprise the following:
1. Host plant research
Mechanism of mlo resistance and pleiotropic phenotypes
Loss of MLO protein function leads to broad-spectrum resistance against
powdery mildew. Our research focuses on identifying the underlying
mechanism. In barley and Arabidopsis, the mlo mutations are
accompanied by negative side effects, so-called pleiotropic phenotypes.
Although those phenotypes have been first described over 30 years ago,
their molecular basis remains unknown. We work on unravelling the
mechanism behind those unwanted side-effects.
2. Barley–B. hordei interaction
Cross-kingdom communication and effector biology
Plants and microbes exchange different types of molecules to
manipulate cellular processes in the interacting organism. We study the
exchange of small non-coding RNAs, such as miRNAs and rRNA- or
tRNA-fragments in the barley-B. hordeiB. hordei and work on
revealing interacting proteins and their mode of action in the barley-B.
hordei interaction and further explore the role of extracellular
vesicles in their transport. Moreover, we are interested in the effector
repertoire of B. hordei and work on revealing interacting
proteins and their mode of action in barley.
3. Arabidopsis–E. cruciferarum interaction
MLO and EXO70 interplay in focal secretion
Some isoforms of membrane-resident MLO proteins confer resistance to the
fungal powdery mildew disease, whereas EXO70 proteins are subunits of
the exocyst complex, which is involved in exocytic vesicle tethering. We
found an isoform-preferential interaction of MLO and EXO70 proteins.
Based on this interaction, we are investigating how MLO and EXO70
proteins interplay in focal secretion.
4. Powdery mildew research
Powdery mildew genomics
Our research focuses on understanding how fungal powdery mildew pathogens
co-evolve with their host plants. We use advanced technologies to
explore the role of transposable elements in powdery mildew evolution,
their impact on fungal genomes, and how they are regulated to prevent
genome damage. One of our key findings was that transposable elements
are induced at specific infection stages, and that they give rise to
novel long noncoding RNAs. We are uncovering how these long noncoding
RNAs contribute to transposon regulation and gene invention in B.
hordei.
Useful links
A general introduction to the
Arabidopsis-powdery mildew interaction can be found in
The Arabidopsis Book.
A recent update of this chapter can be found
here.
|

Figure 1. Barley mlo mutants are broad-spectrum resistant to powdery
mildew. Phenotype of a susceptible ( Mlo , left) and a mlo -resistant
(right) barley genotype inoculated with Blumeria graminis f. sp. hordei K1
conidia and photographed at six days after inoculation.
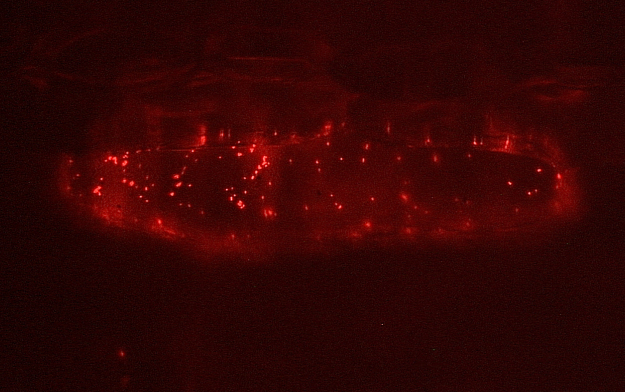
Figure 2. Epifluorescence
micrograph of a barley cell expressing a DsRED-labelled peroxisomal
protein. The bright little dots inside the cell mark (mobile)
peroxisomes.
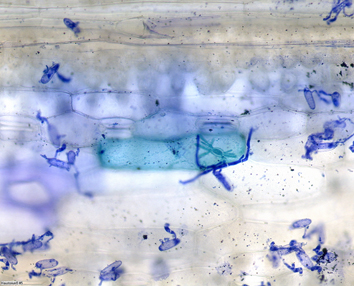
Figure 3. Cell-autonomous
complementation of mlo resistance by transient expression of a Mlo cDNA
in a epidermal cell of a barley mlo genotype. Note the successful
colonization of the transformed beta-glucuronidase
marker gene expressing (greenish) cell as evidenced by the
formation of an intracellular haustorium and hyphal growth.
|